Unsupervised Learning example¶
Nengo Example: Unsupervised learning
When we do error-modulated learning with the nengo.PES rule,
we have a pretty clear idea of what we want to happen.
Years of neuroscientific experiments have yielded
learning rules explaining how synaptic strengths
change given certain stimulation protocols.
But what do these learning rules actually do
to the information transmitted across an
ensemble-to-ensemble connection?
We can investigate this in Nengo.
Currently, we've implemented the nengo.BCM
and nengo.Oja rules.
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import nengo
%load_ext nengo.ipynb
print(nengo.BCM.__doc__)
print(nengo.Oja.__doc__)
Step 1: Create a simple communication channel
The only difference between this network and most models you've seen so far is that we're going to set the decoder solver in the communication channel to generate a full connection weight matrix which we can then learn using typical delta learning rules.
model = nengo.Network()
with model:
sin = nengo.Node(lambda t: np.sin(t*4))
pre = nengo.Ensemble(100, dimensions=1)
post = nengo.Ensemble(100, dimensions=1)
nengo.Connection(sin, pre)
conn = nengo.Connection(pre, post, solver=nengo.solvers.LstsqL2(weights=True))
pre_p = nengo.Probe(pre, synapse=0.01)
post_p = nengo.Probe(post, synapse=0.01)
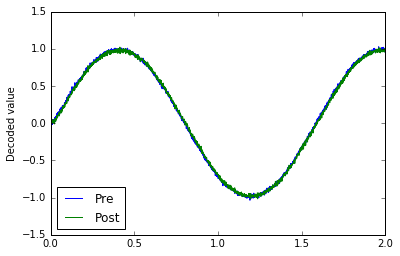
# Verify that it does a communication channel
with nengo.Simulator(model) as sim:
sim.run(2.0)
plt.plot(sim.trange(), sim.data[pre_p], label="Pre")
plt.plot(sim.trange(), sim.data[post_p], label="Post")
plt.ylabel("Decoded value")
plt.legend(loc="best");
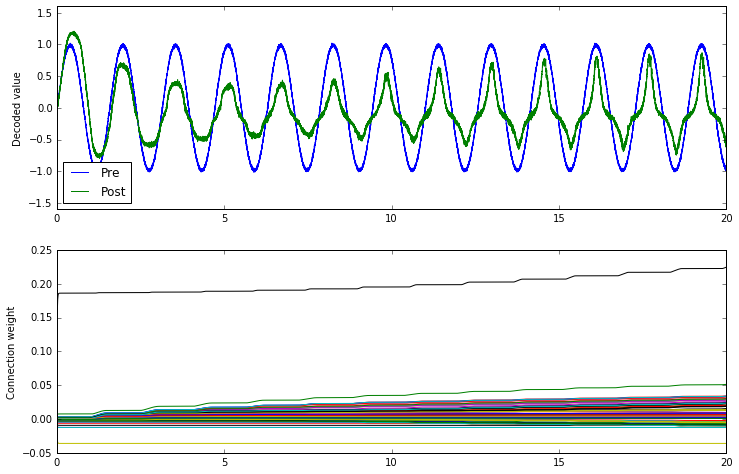
What does BCM do?
conn.learning_rule_type = nengo.BCM(learning_rate=5e-10)
with model:
weights_p = nengo.Probe(conn, 'weights', synapse=0.01, sample_every=0.01)
with nengo.Simulator(model) as sim:
sim.run(20.0)
plt.figure(figsize=(12, 8))
plt.subplot(2, 1, 1)
plt.plot(sim.trange(), sim.data[pre_p], label="Pre")
plt.plot(sim.trange(), sim.data[post_p], label="Post")
plt.ylabel("Decoded value")
plt.ylim(-1.6, 1.6)
plt.legend(loc="lower left")
plt.subplot(2, 1, 2)
# Find weight row with max variance
neuron = np.argmax(np.mean(np.var(sim.data[weights_p], axis=0), axis=1))
plt.plot(sim.trange(dt=0.01), sim.data[weights_p][..., neuron])
plt.ylabel("Connection weight");
The BCM rule appears to cause the ensemble to either be on or off. This seems consistent with the idea that it potentiates active synapses, and depresses non-active synapses.
As well, we can show that BCM sparsifies the weights. The sparsity measure below uses the Gini measure of sparsity, for reasons explained in this paper.
def sparsity_measure(vector): # Gini index
# Max sparsity = 1 (single 1 in the vector)
v = np.sort(np.abs(vector))
n = v.shape[0]
k = np.arange(n) + 1
l1norm = np.sum(v)
summation = np.sum((v / l1norm) * ((n - k + 0.5) / n))
return 1 - 2 * summation
print("Starting sparsity: {0}".format(sparsity_measure(sim.data[weights_p][0])))
print("Ending sparsity: {0}".format(sparsity_measure(sim.data[weights_p][-1])))
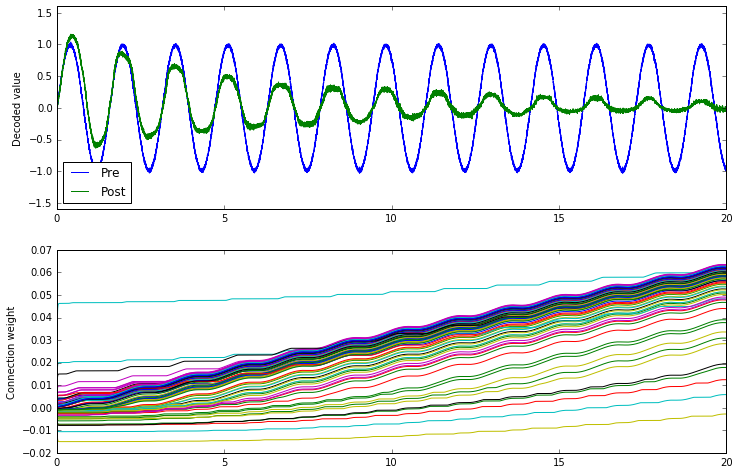
What does Oja do?
conn.learning_rule_type = nengo.Oja(learning_rate=6e-8)
with nengo.Simulator(model) as sim:
sim.run(20.0)
plt.figure(figsize=(12, 8))
plt.subplot(2, 1, 1)
plt.plot(sim.trange(), sim.data[pre_p], label="Pre")
plt.plot(sim.trange(), sim.data[post_p], label="Post")
plt.ylabel("Decoded value")
plt.ylim(-1.6, 1.6)
plt.legend(loc="lower left")
plt.subplot(2, 1, 2)
# Find weight row with max variance
neuron = np.argmax(np.mean(np.var(sim.data[weights_p], axis=0), axis=1))
plt.plot(sim.trange(dt=0.01), sim.data[weights_p][..., neuron])
plt.ylabel("Connection weight")
print("Starting sparsity: {0}".format(sparsity_measure(sim.data[weights_p][0])))
print("Ending sparsity: {0}".format(sparsity_measure(sim.data[weights_p][-1])))
The Oja rule seems to attenuate the signal across the connection.
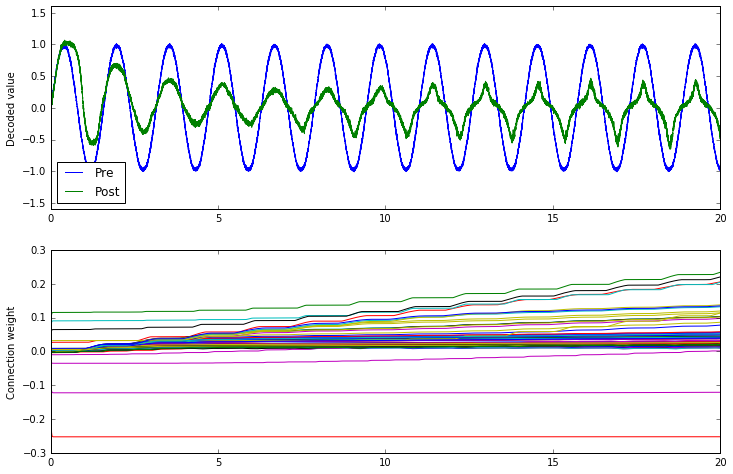
What do BCM and Oja together do?
We can apply both learning rules to the same connection
by passing a list to learning_rule_type.
conn.learning_rule_type = [nengo.BCM(learning_rate=5e-10), nengo.Oja(learning_rate=2e-9)]
with nengo.Simulator(model) as sim:
sim.run(20.0)
plt.figure(figsize=(12, 8))
plt.subplot(2, 1, 1)
plt.plot(sim.trange(), sim.data[pre_p], label="Pre")
plt.plot(sim.trange(), sim.data[post_p], label="Post")
plt.ylabel("Decoded value")
plt.ylim(-1.6, 1.6)
plt.legend(loc="lower left")
plt.subplot(2, 1, 2)
# Find weight row with max variance
neuron = np.argmax(np.mean(np.var(sim.data[weights_p], axis=0), axis=1))
plt.plot(sim.trange(dt=0.01), sim.data[weights_p][..., neuron])
plt.ylabel("Connection weight")
print("Starting sparsity: {0}".format(sparsity_measure(sim.data[weights_p][0])))
print("Ending sparsity: {0}".format(sparsity_measure(sim.data[weights_p][-1])))
The combination of the two appears to accentuate the on-off nature of the BCM rule. As the Oja rule enforces a soft upper or lower bound for the connection weight, the combination of both rules may be more stable than BCM alone.
That's mostly conjecture, however; a thorough investigation of the BCM and Oja rules and how they interact has not yet been done.
Download learn_unsupervised as an IPython notebook or Python script.