Advanced network design example¶
Additional tips and tricks for designing networks
This tutorial assumes that you have read
the network_design tutorial,
and have designed a network or two.
Here, we will give a few advanced tips and tricks
for designing networks that can be reused flexibly.
In particular, these tips will use the
config system, so we will also assume that
you have gone over the config tutorial.
Briefly, the general principles covered in this tutorial are
- Accept a network argument
- Accept a config argument for groups of parameters
We will demonstrate these principles
using the two examples from the network_design tutorial.
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import nengo
%load_ext nengo.ipynb
from nengo.dists import Choice
from nengo.utils.functions import piecewise
from nengo.utils.ipython import hide_input
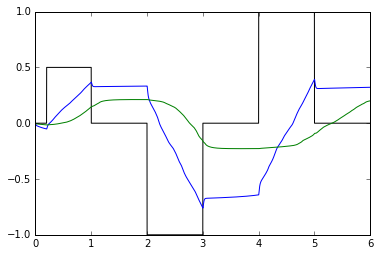
def test_integrators(net):
with net:
piecewise_f = piecewise({0: 0, 0.2: 0.5, 1: 0, 2: -1, 3: 0, 4: 1, 5: 0})
piecewise_inp = nengo.Node(piecewise_f)
nengo.Connection(piecewise_inp, net.pre_integrator.input)
input_probe = nengo.Probe(piecewise_inp)
pre_probe = nengo.Probe(net.pre_integrator.ensemble, synapse=0.01)
post_probe = nengo.Probe(net.post_integrator.ensemble, synapse=0.01)
with nengo.Simulator(net) as sim:
sim.run(6)
plt.plot(sim.trange(), sim.data[input_probe], color='k')
plt.plot(sim.trange(), sim.data[pre_probe], color='b')
plt.plot(sim.trange(), sim.data[post_probe], color='g')
hide_input()
1. Accept a network argument
Typically, a network-creation function
will take a set of arguments,
which affect some important parts
of the network.
When testing the network,
it's common to change several parameters
to see how they affect the network.
One way to do this is to add more and more
arguments to your function;
this quickly gets out of hand.
Instead, use the config system,
which enables us to set
network-level defaults for all Nengo objects.
You can either do this by creating your network in the context of some other network, or you can modify your function to optionally take in a network instance, which you will build your objects into.
In the example below,
we change both integrators to use LIFRate neurons
by changing the config object
in the network both integrators
are build within.
We also change the post_integrator
to use a very small radius
by passing in a network
which has its default radius modified.
def Integrator(n_neurons, dimensions, tau=0.1, net=None):
if net is None:
net = nengo.Network()
with net:
net.input = nengo.Node(size_in=dimensions)
net.ensemble = nengo.Ensemble(n_neurons, dimensions=dimensions)
nengo.Connection(net.ensemble, net.ensemble, synapse=tau)
nengo.Connection(net.input, net.ensemble,
synapse=None, transform=tau)
return net
net = nengo.Network(label="Two integrators")
with net:
# Make both integrators use LIFRate neurons
net.config[nengo.Ensemble].neuron_type = nengo.LIFRate()
net.pre_integrator = Integrator(50, 1)
# Lower the radius of the post_integrator
net.post_integrator = nengo.Network()
net.post_integrator.config[nengo.Ensemble].radius = 0.2
Integrator(50, 1, net=net.post_integrator)
nengo.Connection(net.pre_integrator.ensemble, net.post_integrator.input)
test_integrators(net)
2. Accept a config argument for groups of parameters
Often, you will not want to use the
network-level defaults for all of your objects.
Some objects need certain things overwritten,
while others need other values overwritten.
Again, it is possible to deal with this issue
by adding more and more parameters,
but this quickly gets out of hand.
Instead, add a small number of arguments
that optionally accept a config object,
which allows for setting multiple parameters at once.
In the coupled integrator network example,
we make two connections.
We have to be careful changing the defaults
for those connections, as they are wildly different;
one is a recurrent connection from an ensemble to itself,
while the other is a connection from a node to an ensemble.
We will accept a config object for the recurrent connection
to make this easier.
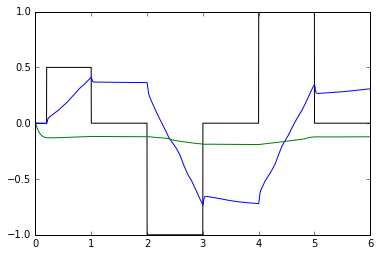
def Integrator(n_neurons, dimensions, recurrent_config=None, net=None):
if net is None:
net = nengo.Network()
if recurrent_config is None:
recurrent_config = nengo.Config(nengo.Connection)
recurrent_config[nengo.Connection].synapse = nengo.Lowpass(0.1)
with net:
net.input = nengo.Node(size_in=dimensions)
net.ensemble = nengo.Ensemble(n_neurons, dimensions=dimensions)
with recurrent_config:
nengo.Connection(net.ensemble, net.ensemble)
tau = nengo.Config.default(nengo.Connection, 'synapse').tau
nengo.Connection(net.input, net.ensemble,
synapse=None, transform=tau)
return net
net = nengo.Network(label="Two integrators")
with net:
# Make both integrators use LIFRate neurons
net.config[nengo.Ensemble].neuron_type = nengo.LIFRate()
net.pre_integrator = Integrator(50, 1)
# Give the post_integrator a shorter tau (should make integration fail)
recurrent_config = nengo.Config(nengo.Connection)
recurrent_config[nengo.Connection].synapse = nengo.Lowpass(0.01)
net.post_integrator = Integrator(50, 1, recurrent_config=recurrent_config)
nengo.Connection(net.pre_integrator.ensemble, net.post_integrator.input)
test_integrators(net)
Longer example: double integrator network
Recall in the previous tutorial that
we created a model
that released a lever 0.6 to 1.0 seconds
after pressing a lever.
Let's use the above principles,
and the config system in general,
to improve the code constructing this model.
def controlled_integrator(n_neurons, dimensions, recurrent_config=None, net=None):
if net is None:
net = nengo.Network()
if recurrent_config is None:
recurrent_config = nengo.Config(nengo.Connection)
recurrent_config[nengo.Connection].synapse = nengo.Lowpass(0.1)
with net:
net.ensemble = nengo.Ensemble(n_neurons, dimensions=dimensions + 1)
with recurrent_config:
nengo.Connection(net.ensemble, net.ensemble[:dimensions],
function=lambda x: x[:-1] * (1.0 - x[-1]))
return net
def medial_pfc(coupling_strength, n_neurons_per_integrator=200, recurrent_config=None, tau=0.1, net=None):
if net is None:
net = nengo.Network()
with net:
recurrent_config = nengo.Config(nengo.Connection)
recurrent_config[nengo.Connection].synapse = nengo.Lowpass(tau)
net.pre = controlled_integrator(n_neurons_per_integrator, 1, recurrent_config)
net.post = controlled_integrator(n_neurons_per_integrator, 1, recurrent_config)
nengo.Connection(net.pre.ensemble[0], net.post.ensemble[0],
transform=coupling_strength)
return net
def motor_cortex(command_threshold, n_neurons_per_command=30, ens_config=None, net=None):
if net is None:
net = nengo.Network()
if ens_config is None:
ens_config = nengo.Config(nengo.Ensemble)
ens_config[nengo.Ensemble].encoders = Choice([[1]])
ens_config[nengo.Ensemble].intercepts = Choice([command_threshold])
with net:
with ens_config:
net.press = nengo.Ensemble(n_neurons_per_command, dimensions=1)
net.release = nengo.Ensemble(n_neurons_per_command, dimensions=1)
return net
def double_integrator(mpfc_coupling_strength,
command_threshold,
press_to_pre_gain=3,
press_to_post_control=-6,
recurrent_tau=0.1,
net=None):
if net is None:
net = nengo.Network()
with net:
net.mpfc = medial_pfc(mpfc_coupling_strength)
net.motor = motor_cortex(command_threshold)
nengo.Connection(net.motor.press, net.mpfc.pre.ensemble[0],
transform=recurrent_tau * press_to_pre_gain)
nengo.Connection(net.motor.press, net.mpfc.post.ensemble[1],
transform=press_to_post_control)
nengo.Connection(net.mpfc.post.ensemble[0], net.motor.release)
return net
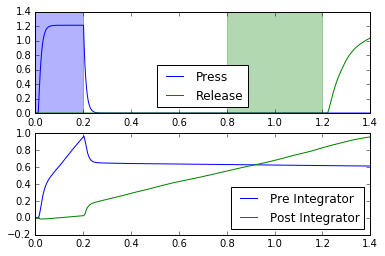
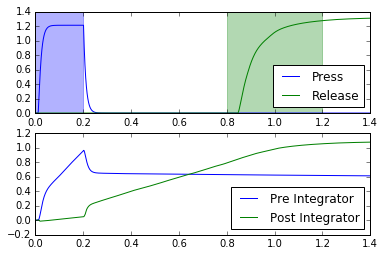
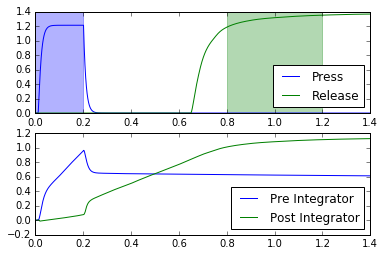
def test_doubleintegrator(net):
# Provide input and probe outside of network construction, for more flexibility
with net:
nengo.Connection(nengo.Node(lambda t: 1 if t < 0.2 else 0), net.motor.press)
pr_press = nengo.Probe(net.motor.press, synapse=0.01)
pr_release = nengo.Probe(net.motor.release, synapse=0.01)
pr_pre_int = nengo.Probe(net.mpfc.pre.ensemble[0], synapse=0.01)
pr_post_int = nengo.Probe(net.mpfc.post.ensemble[0], synapse=0.01)
with nengo.Simulator(net) as sim:
sim.run(1.4)
t = sim.trange()
plt.figure()
plt.subplot(2, 1, 1)
plt.plot(t, sim.data[pr_press], c='b', label="Press")
plt.plot(t, sim.data[pr_release], c='g', label="Release")
plt.axvspan(0, 0.2, color='b', alpha=0.3)
plt.axvspan(0.8, 1.2, color='g', alpha=0.3)
plt.xlim(right=1.4)
plt.legend(loc="best")
plt.subplot(2, 1, 2)
plt.plot(t, sim.data[pr_pre_int], label="Pre Integrator")
plt.plot(t, sim.data[pr_post_int], label="Post Integrator")
plt.xlim(right=1.4)
plt.legend(loc="best")
for coupling_strength in (0.11, 0.16, 0.21):
net = nengo.Network(seed=0) # Set seed here instead
# Try the same network with LIFRate neurons
net.config[nengo.Ensemble].neuron_type = nengo.LIFRate()
net = double_integrator(mpfc_coupling_strength=coupling_strength,
command_threshold=0.85,
net = net)
test_doubleintegrator(net)
Download network_design_advanced as an IPython notebook or Python script.